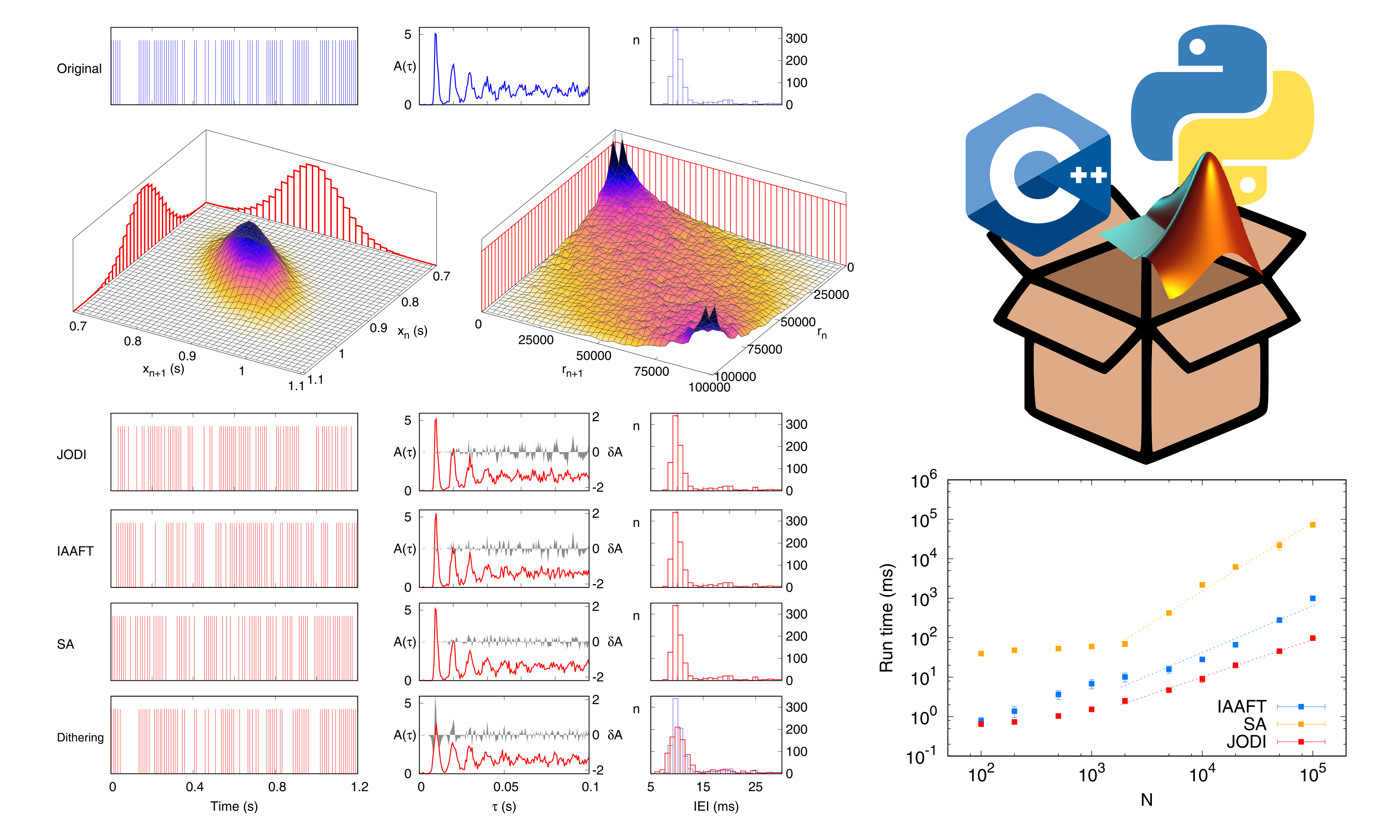
The study of many dynamical systems relies on the analysis of experimentally-recorded time series of events for which information is encoded in the sequence of inter-event intervals. A correct interpretation of the results of the application of analytical techniques to these sequences requires the assessment of statistical significance. In most cases, the corresponding null-hypothesis distribution is unknown, thus forbidding an evaluation of the significance. An alternative solution, which is efficient in the case of continuous signals, is provided by the generation of surrogate data that share statistical and spectral properties with the original dataset. However, in the case of event sequences, the available algorithms for the generation of surrogate data can become cumbersome and computationally demanding. We developed a new method, called "JODI", for the generation of surrogate event sequences that relies on the joint distribution of successive interevent intervals. Our method, which was tested on both synthetic and experimental sequences, performs equally well or even better than conventional methods in terms of interevent interval distribution and autocorrelation while abating the computational time by at least one order of magnitude. The method was first published in:
L. Ricci, M. Castelluzzo, L. Minati and A. Perinelli, Generation of surrogate event sequences via joint distribution of successive inter-event intervals, Chaos 29 (2019), 121102, doi:10.1063/1.5138250
In addition, we developed a software package that implements four algorithms for the generation of surrogate data out of spike trains and more generally out of any sequence of discrete events. Besides JODI, the package also implements three among the most used surrogate generation algorithms, namely IAAFT, simulated annealing, and dithering. The purpose of the package is to provide a unified and portable toolbox to carry out surrogate generation on point-process data. Code is provided in three languages, namely, C++, Matlab, and Python, thus allowing straightforward integration of package functions into most analysis pipelines. The SpiSeMe (Spike Sequence Mime) package, available in a dedicated GitHub repository, is thoroughly described in:
A. Perinelli, M. Castelluzzo, L. Minati and L. Ricci, SpiSeMe: A multi-language package for spike train surrogate generation, Chaos 30 (2020), 073120, doi: 10.1063/5.0011328